1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
| input_file = "./data/retroplume_raw/traj/clustered_ind/2018-03-30-example.txt"
with open(input_file, "r") as file:
lines = file.readlines()[7:]
data = [list(map(float, line.split())) for line in lines]
df_traj = pd.DataFrame(data, columns=columns[:len(data[0])])
kp_traj = df_traj[df_traj['receptor'] == 1.0].reset_index(drop = True)
dl_traj = df_traj[df_traj['receptor'] == 2.0].reset_index(drop = True)
trajectories = {
"KP": {
"lon": kp_traj['meanLon'],
"lat": kp_traj['meanLat'],
"alt": kp_traj['meanZ']/1000.0 + kp_traj['meanTopo']/1000.0,
"time_delta": -1*(kp_traj['time'] - kp_traj['time'].iloc[0])/60/60.0,
},
"DL": {
"lon": dl_traj['meanLon'],
"lat": dl_traj['meanLat'],
"alt": dl_traj['meanZ']/1000.0 + dl_traj['meanTopo']/1000.0,
'time_delta':-1*(kp_traj['time'] - kp_traj['time'].iloc[0])/60/60.0
},
}
elevation_interpolator = RegularGridInterpolator(
(lat, lon), elevation / 1000.0
)
def set_3d_background(ax,sel_df_bio):
ax.scatter(
80.331871, 26.449923, 0.35,
zdir="z",
marker="s",
facecolor='none',
edgecolor="b",
s=40,
alpha=1,lw = 2,
label="Kanpur",zorder = 3
)
ax.scatter(
77.069710, 28.679079, 0.53,
zdir="z",
marker="s",
facecolor='none',
edgecolor="r",
s=40,
alpha=1,lw = 2,
label="New Delhi",zorder = 3
)
ax.plot_surface(
lon_grid,
lat_grid,
subset_elevation/1000.0,
cmap=plt.cm.gray,
rstride=10,
cstride=10,
edgecolor='none',
alpha=0.15,
)
ax.xaxis._axinfo["grid"]["color"] = (1, 1, 1, 0)
ax.yaxis._axinfo["grid"]["color"] = (1, 1, 1, 0)
ax.zaxis._axinfo["grid"]["color"] = (1, 1, 1, 0)
ax.set_xlim([lon_min,lon_max])
ax.set_ylim([lat_min, lat_max])
ax.set_xlabel("Longitude")
ax.set_ylabel("Latitude")
ax.set_zlabel("Altitude (km)")
def plot_individual_day_traj(ax,traj):
norm = mcolors.Normalize(vmin=0, vmax=72)
cmap = cm.get_cmap('Spectral_r')
for i in range(len(traj["time_delta"]) - 1):
ax.plot(
traj["lon"][i:i+2],
traj["lat"][i:i+2],
traj["alt"][i:i+2],
color=cmap(norm(traj["time_delta"][i])),
linewidth=2.5,zorder =15,
)
fig = plt.figure(figsize=(6, 5))
norm = mcolors.Normalize(vmin=0, vmax=72)
cmap = cm.get_cmap('Spectral_r')
ax = fig.add_subplot(111, projection='3d')
plot_individual_day_traj(ax, trajectories['DL'])
set_3d_background_dl(ax,sel_df_bio)
ax.set_zlim([0, 7])
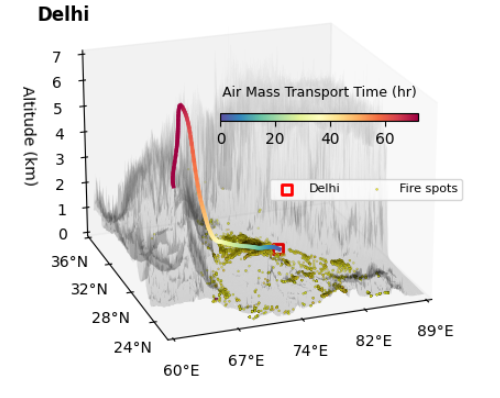
ax.set_title('Delhi', loc = 'left', fontweight = 'bold',y=0.85)
ax.legend(ncol = 3, loc = (0.55, 0.45), fontsize = 8, frameon=True)
ax.view_init(elev=21, azim=-110)
plt.show()
|
Comments