1. Motivation
Continuing from the previous post on molecular formula assignment, I encountered a common issue: overfitting during TOF peak assignment. In some cases, a narrow TOF m/z window was being assigned with too many candidate formulas, resulting in high uncertainty for each individual ion. In other words, we might obtain a wrong intensity for a correct formula, which affecting the dat reliability.
To address this, I implemented a refinement step after formula assignment:
Only the local maxima within a given ppm range are retained, along with known TOF apex peaks.
2. Concept
The idea is simple:
- If multiple peaks are detected within a small ppm window, keep only the most intense one.
- If a lower-intensity peak is near a known TOF apex position (from missing peaks), it is also retained to preserve chemically real signals that Orbitrap may have only partially captured. (this is because sometimes there are multiple peaks in an unit m/z window, and there are some ion on the shoulder of the main peak, and it is close to the small peak apex, if they somehow matched with the previous criteria, the ion on the peak apex were removed,which was not we expced.)
3. Function of Intensity-Based Refiner
1 | def refiner_by_orbitrap_intensity_ratio(mf_result, ppm_range=50, missing_peak_list=None): |
4. Applying the Refiner
1 | refining_distance = 40 # in ppm |
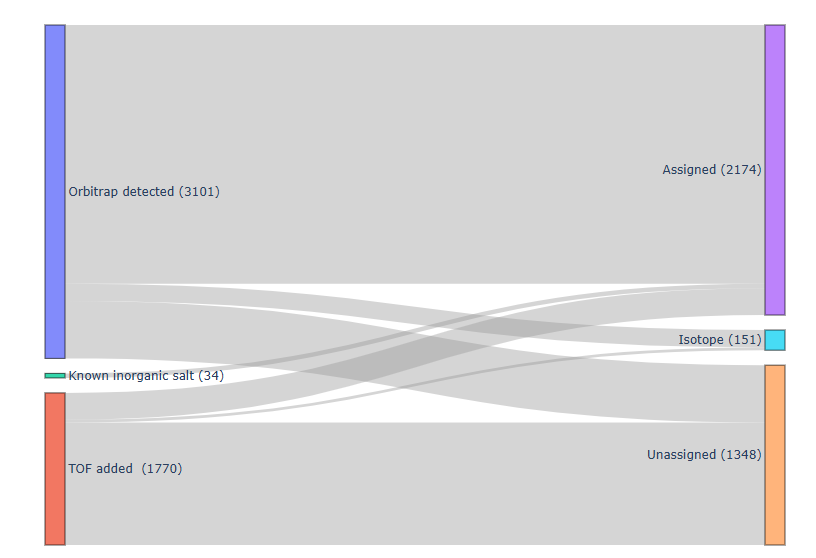
After refinement, the final peak list can be exported in text format for direct use in TOFware fitting. This refined list includes:
- Orbitrap-resolved peaks
- TOF-reconstructed (“missing”) peaks
- Known inorganic salt peaks (primary ions and its clusters, K+, Mg+, etc)
While isotope peaks are excluded from the fitting, unassigned ions are retained. Although these unassigned ions may not provide direct chemical information, their presence helps prevent signal over-allocation to nearby assigned peaks — particularly in dense or overlapping m/z regions.
An illustration of the ion inflow–outflow process is shown below as a Sankey diagram.
Before integrating Orbitrap into the TOF fitting workflow, only ~2000 ions could be manually fit, often with significant chemical ambiguity. With the addition of Orbitrap’s ultra-high-resolution molecular information, the number of confidently assigned ions has increased to over 3500, significantly enhancing the chemical characterization of the analytes.
Comments